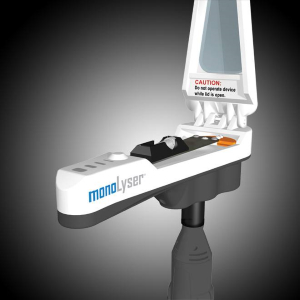
MonoLyser™ the World’s Fastest Lab and Field Life Science Homogenizer
The MonoLyser™ is the world’s fastest Portable Lab and Field Life Science Homogenizer. This is an ultrafast lysing tool that can be used in the field or in a laboratory. This innovative device incorporates RotaPrep’s Hyperlysing technology and lyse most samples in a matter of seconds. The complete homogenization lab includes: the MonoLyser, rotary drive tool, two batteries, and charger.
Too often in today’s molecular biology labs the sample lysis is a bottleneck which delays test results and requires human interaction and follow up for the entire process. The MonoLyser™ does away with that!
The MonoLyser™ system is a sample lysis, homogenization and grinding system for life sciences applications like no other on the market. It is for an order of magnitude faster in performing complete quantitative sample lysis, usually within 5 seconds or less, of even the most difficult biological and environmental samples. You can process any sample for DNA extraction, RNA extraction, and protein extraction in the field, or in lab with ease. Most samples are completely lysed and ready for downstream processing within the 5 seconds!!! Traditional methods of sample lysis require tedious homogenization using a pestle and mortar, ultrasonication, bench scale bead beating equipment, a shearing homogenizer, or enzymatic or chemical treatments. Those processes run from tens of hours to 40 seconds in processing time and quite often produce a bottleneck in the workflow. It is compatible with any downstream protocol for DNA purification, RNA purification, proteins purification, and metabolites purification and characterization.
The MonoLyser™ represents novel thinking in the area of sample lysis. It is a portable omni-directional bead beating system with a unique, patent pending balanced crankshaft–slider mechanism which creates the most aggressive bead beating lysis based on combination of cascade impaction, shearing, and if the sample is in buffer, shear flow lysis. The combined kinetics of the processes result in an amazing performances of 5 seconds or less lysing time for most samples, something unheard of in the field!
The MonoLyser™ is an attachment that fits on a high speed rotary tool. Attachment to the tool is done via a built-in threaded adapter. Rotary tools are available in DC (battery) powered versions for field use or AC powered versions, which are ideal for continuous use in a laboratory setting. The high end rotary tools come with an on/off switch separate from the speed control, making it easy to run multiple samples at the same speed.
Application: MonoLyser™ Delivers Almost Instantaneous Lysis
The most valuable feature of our system is an almost instantaneous lysis!
The Monolyser’s unique super-fast kinetics of grinding eliminates most of the time spent in the sample preparation bottleneck. If you work in the field, you are not obliged to bring back entire specimens, just the high quality lysate or extracted DNA for analysis.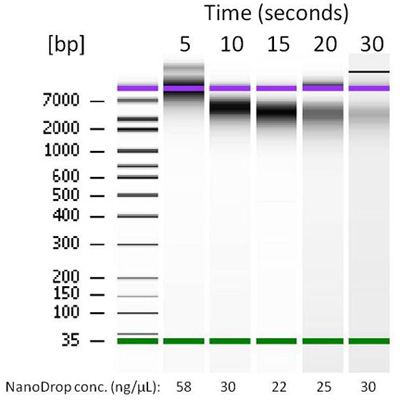
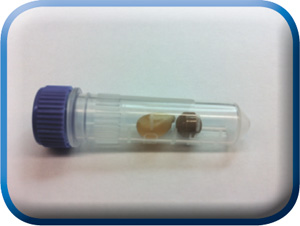
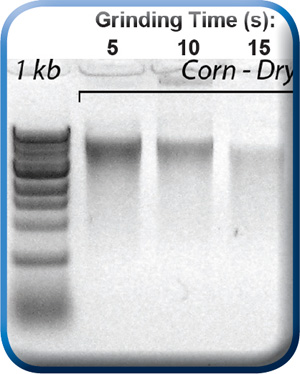
DNA released from corn kernel after 5, 10, 15, 20, and 30 seconds of grinding by the MonoLyser™. Extraction and purification was performed with a Zymo Research ZR-Plant/Seed DNA MiniPrep kit.
Sample in a 2mL, either conical tube filled with bead beating lysing matrix, is placed in the tube holder by inserting it in, and closing the safety door. By activating the tool at 2/3 of maximum speed setting for 5 seconds, holding it by hand or placing it on a laboratory stand, the complete dry or wet grinding of the even most difficult sample is quantitatively performed. The figure above shows the contents of the tube after dry grinding of a dry corn kernel, one of the most difficult agro/bio samples to process. In order to demonstrate the quality of lysate we performed complete DNA purification followed by analysis with a NanoDrop spectrophotometer and a BioAnalyser system from Agilent Technologies. We used an MP Bio Matrix A tube with two Ľ inch ceramic balls for grinding and a Zymo Research Plant/Seed DNA MiniPrep kit for DNA extraction and purification. After lysis, the sample was centrifuged and the supernatant was processed manually through the kit using a bind-wash-elute column based protocol described in the instructions of the kit. The resulting DNA BioAnalyser image is presented above, together with the DNA quantification data. At 5 seconds, the high quality and quantity of extracted DNA with low shearing is evident. At the longer grinding times, increasing amounts of shearing of DNA as well as decreasing average sizes were observed. This is the shortest time from hard and difficult sample to lysate in the industry. Comparable high end benchtop homogenizers perform the same task for approximately 20 – 40 seconds. The quality and quantity of the extracted DNA in 5 second run is comparable to those obtained by the most current lab bench high throughput homogenizers based upon multi-directional bead beating.